IMC F03A Operations on Sequence & Feature Viewer Pane
Display range
- The contents of the display field at the top. This display changes line by line by scrolling.
- Length: Base length currently displayed in the pane (unit: bp)
- From: Absolute starting position on the genome of the base sequence currently displayed in the pane
- To: Absolute termination position on the genome of the base sequence currently displayed in the pane
Display range scrolling
- Click the red arrows on the top left and right.
- Click to shift left or right by one line.
- You will scroll up and down line by line.
Change the number of displayed bases in one line
- You can change it by resizing the width of the Annotation Window.
Change the number of lines displayed in the pane
- You can change it by resizing the height of the Annotation Window.
Minimal display
- Only the base sequence of the forward chain is displayed.
- Various options can be displayed by operating the button tool on the left side.
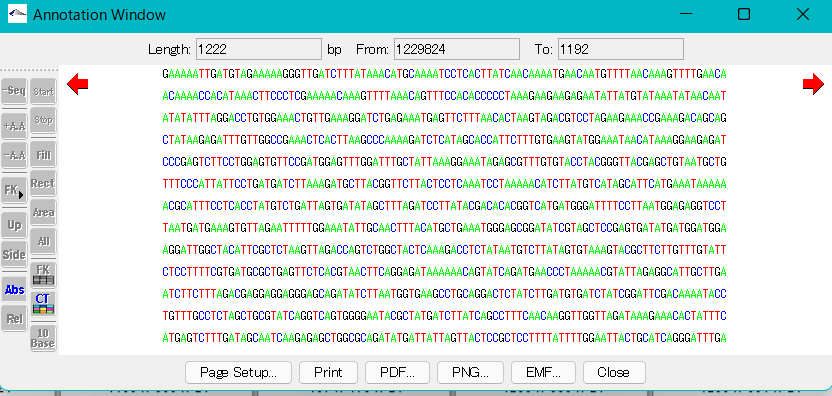
Display of complementary strand base sequence
- Turn on the -Seq button in the left toolbox.
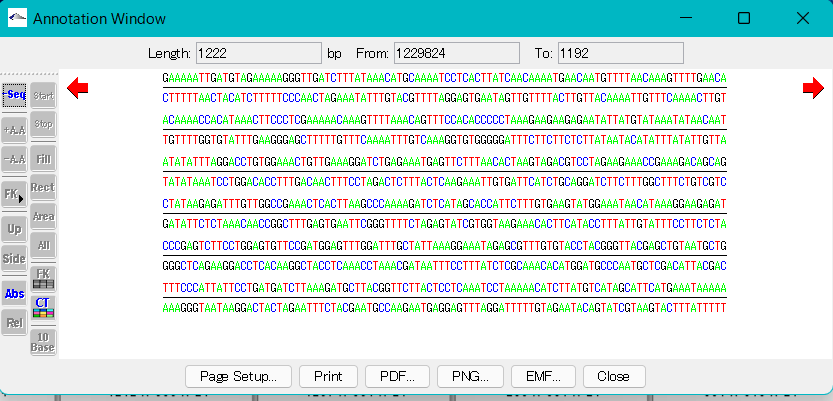
Display of amino acid sequence
- Turn on the + A.A. or -A.A. buttons in the toolbox.
- It is also possible to turn on the two at the same time.
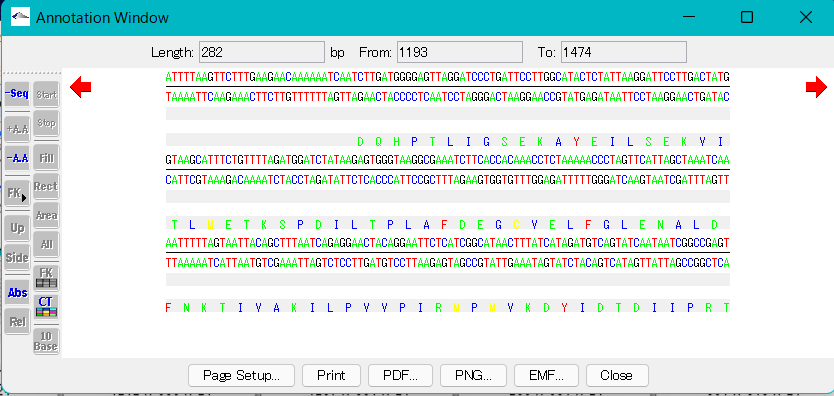
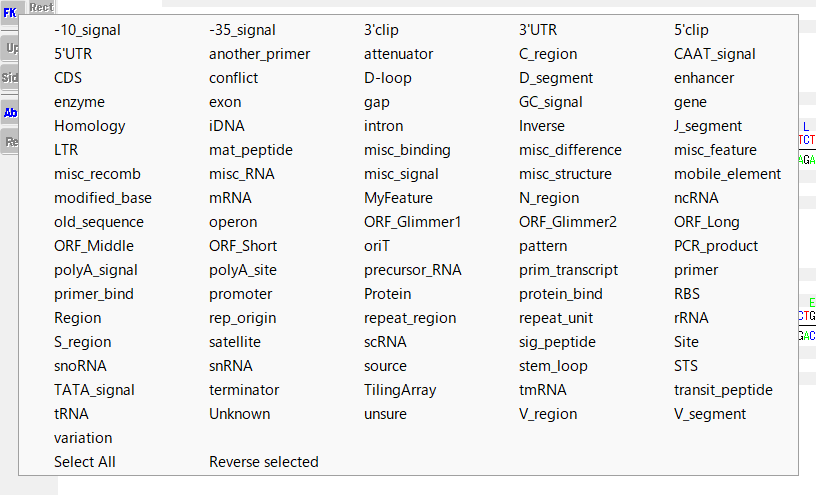
Displaying features
- You can graphically display the selected feature keys above the sequence by turning on the FK button in the toolbox.
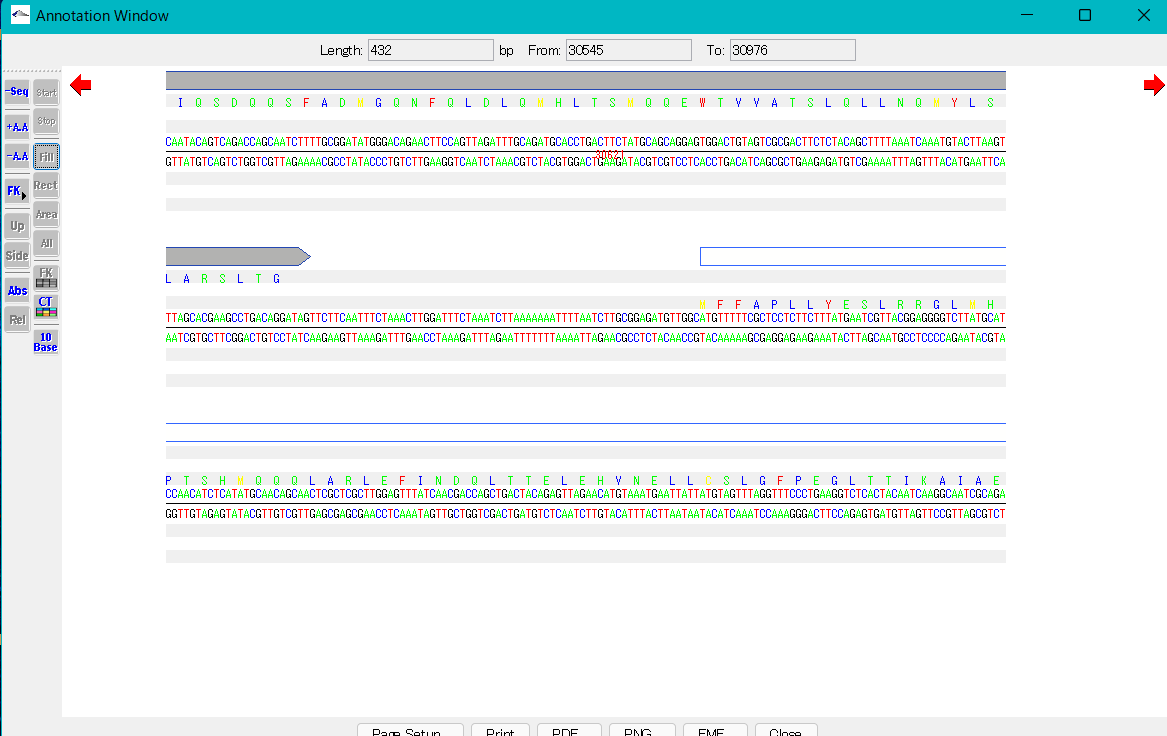
How to select display features
- Right-click on the FK button.
- A pop-up menu will appear showing all feature keys.
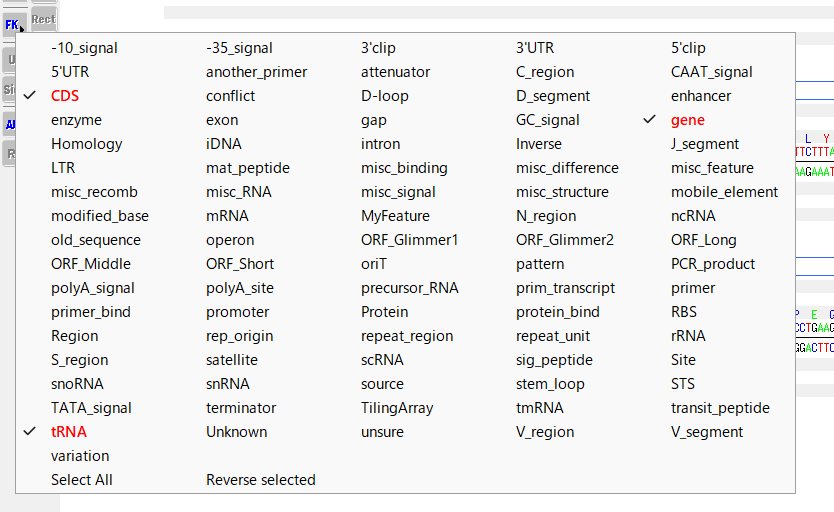
- The feature key displayed in red font is the display target.
- You can change the feature key to be displayed by right-clicking on the feature displayed in black font.
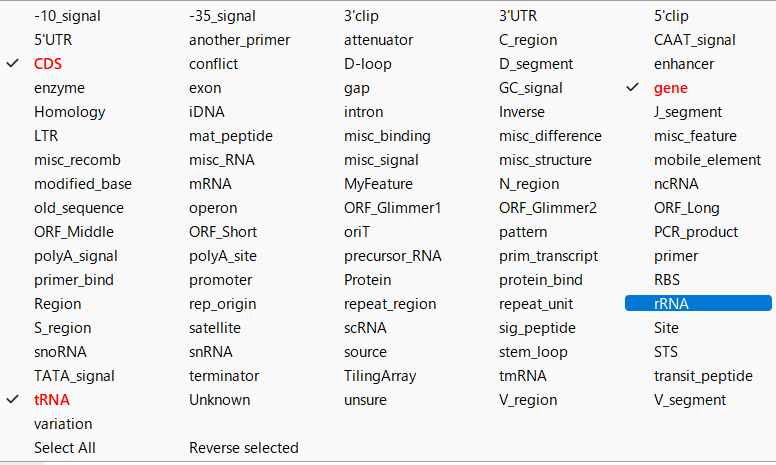
- When changed, the feature key will be displayed in red font.
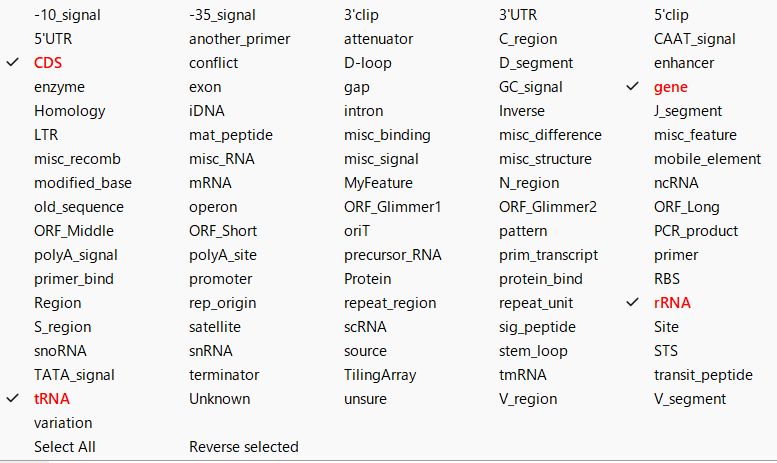
Switching display feature selection status
- Select All: Select all feature keys.
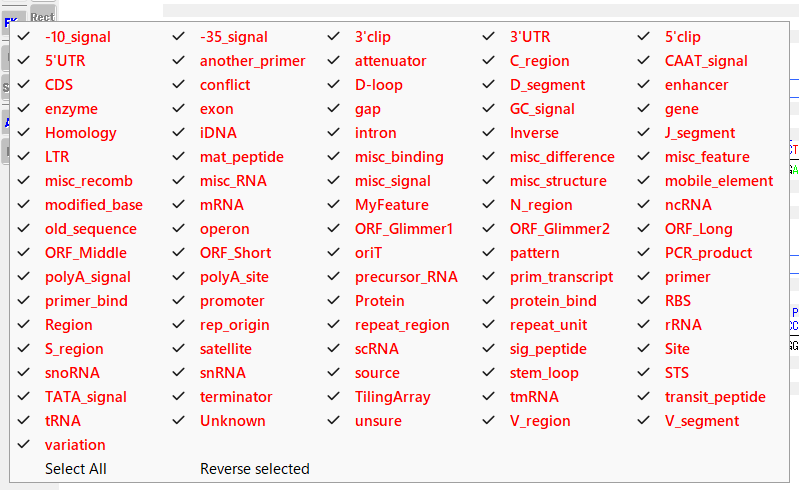
- Reverse Selected: Switches the currently selected feature key to unselected and the unselected feature key to selected all at once.
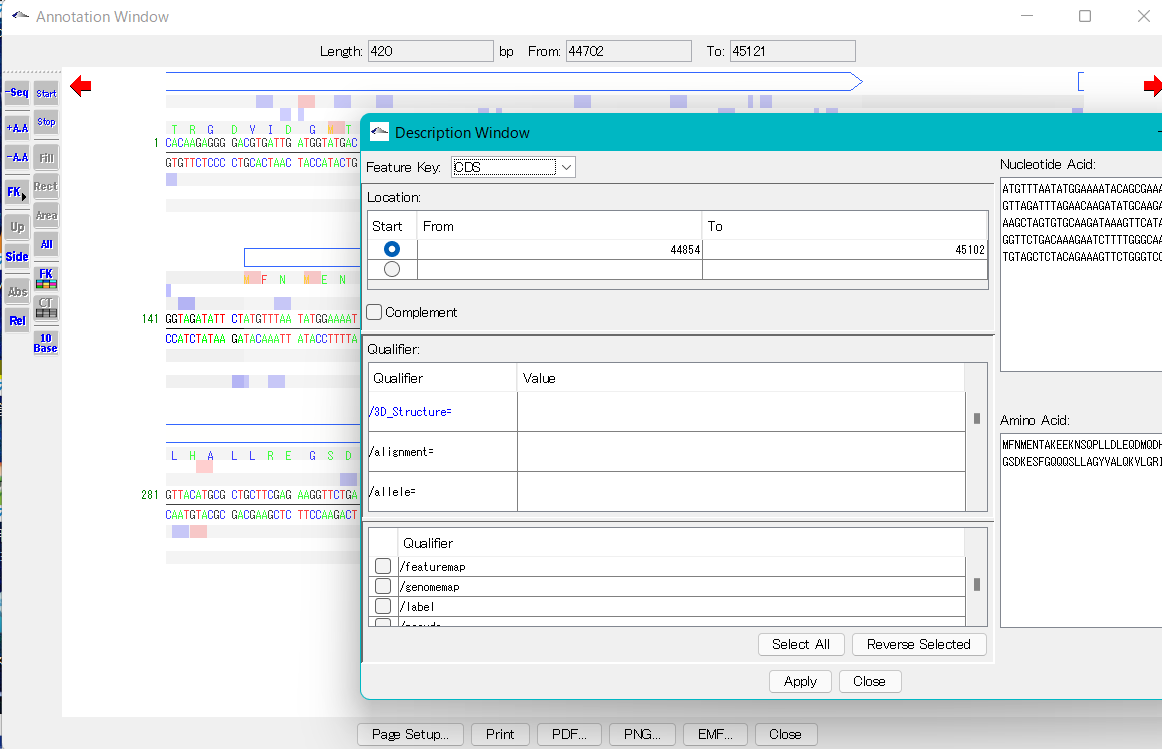
Launching the Description Window
- You can launch the Description Window for a feature by right-clicking on the feature.
- The Description pane of the Annotation Window will be opened in a separate window.
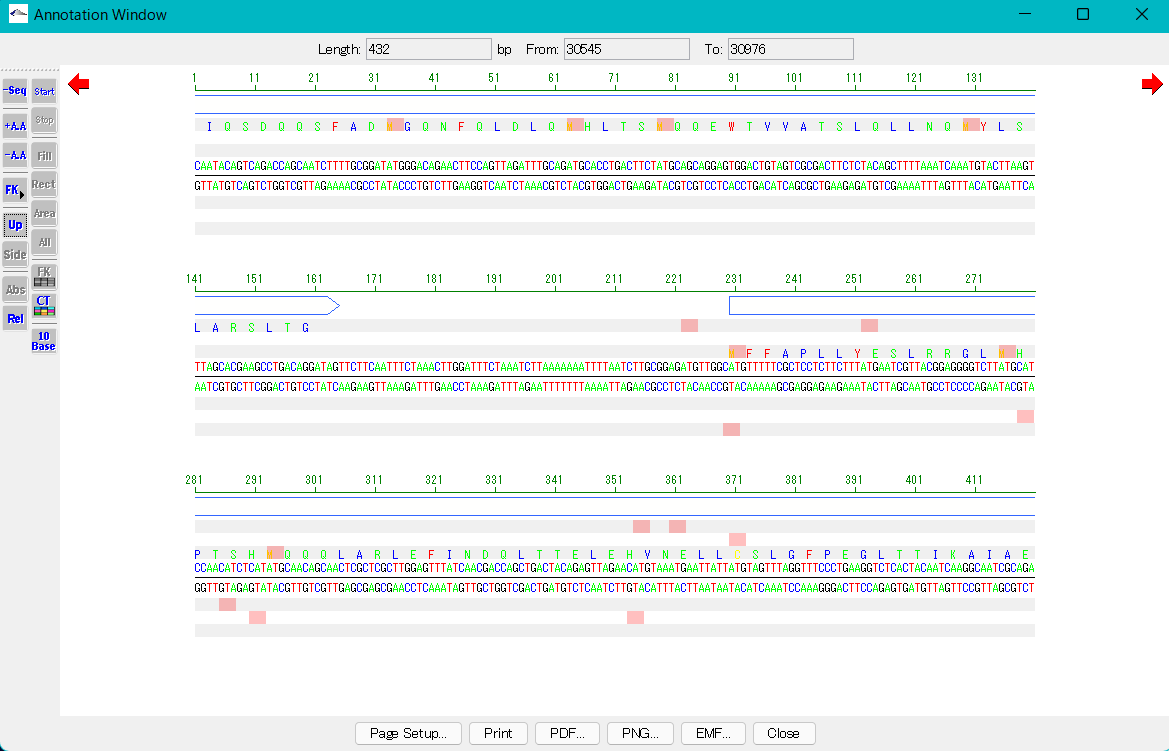
Display of base position scale
- When you turn on the Up button in the toolbox, the position scale of the base sequence is displayed at the top of the sequence.
- When the Side button in the toolbox is turned on, the position scale of the base sequence is displayed on the left side of each row of the sequence.
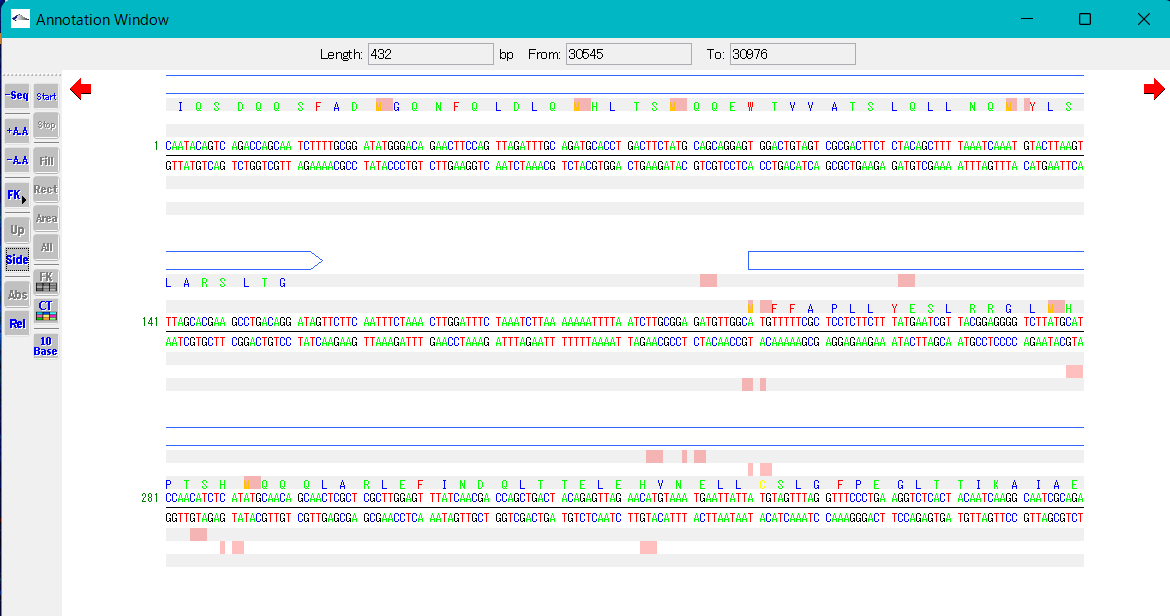
Switching between absolute and relative display of the base sequence scale
- When the Abs button in the toolbox is turned on, the base position scale is displayed at the absolute base position of the loaded current genomic sequence.
- When the Rel button in the toolbox is turned on, the base sequence scale is displayed with the first base in the currently displayed range as 1.
Display of start codon and stop codon candidates
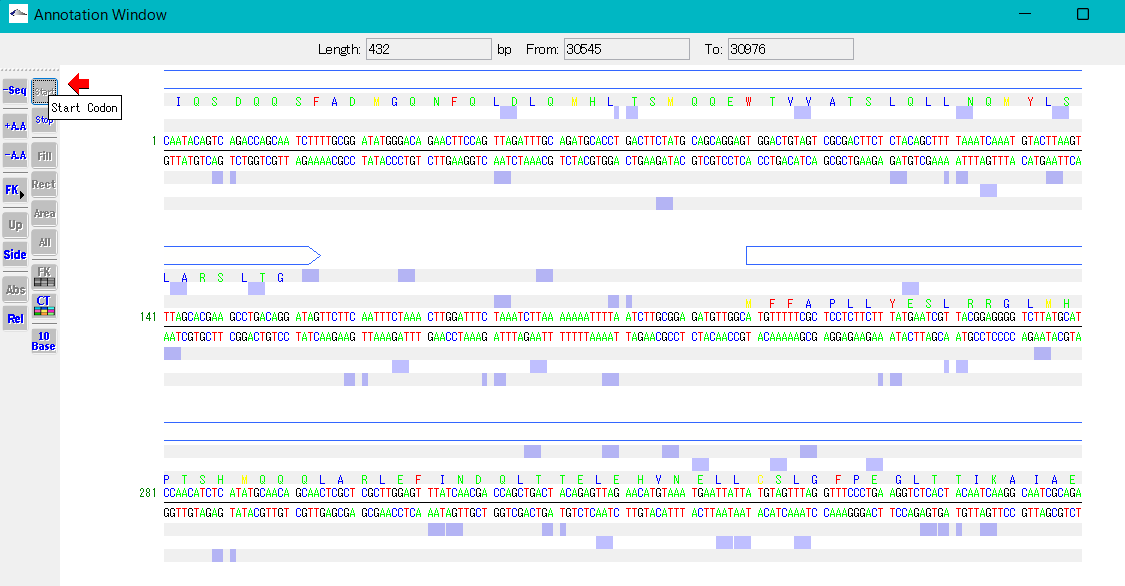
- When you turn on the Start button in the toolbox, the start codon candidate positions are displayed on the base sequence and amino acid display frames.
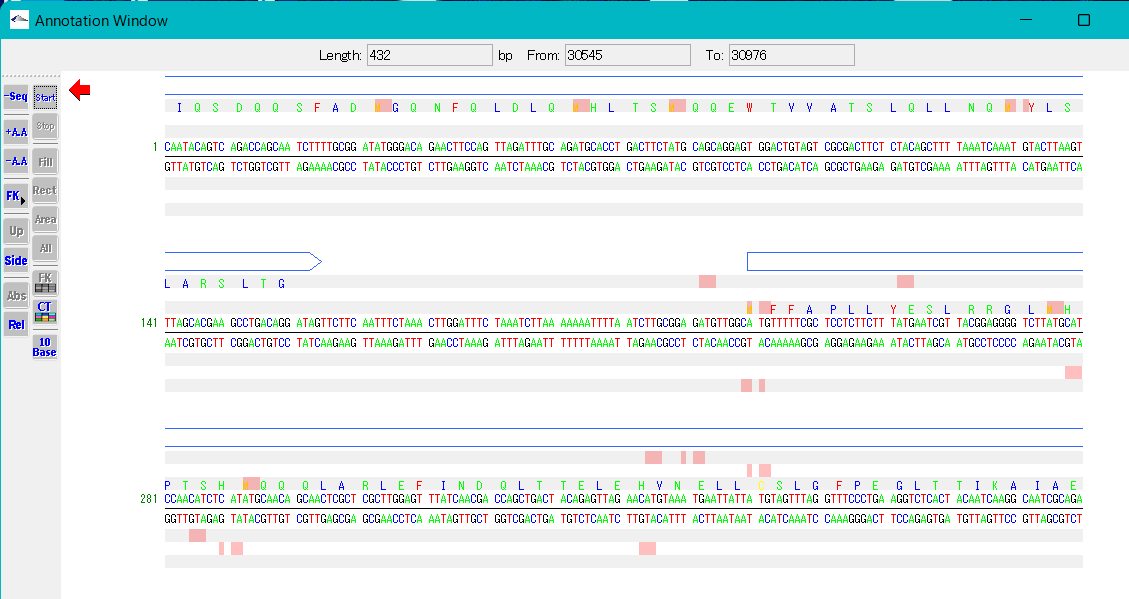
- When you turn on the Stop button in the toolbox, stop codon candidate positions are displayed on the base sequence and amino acid display frames.
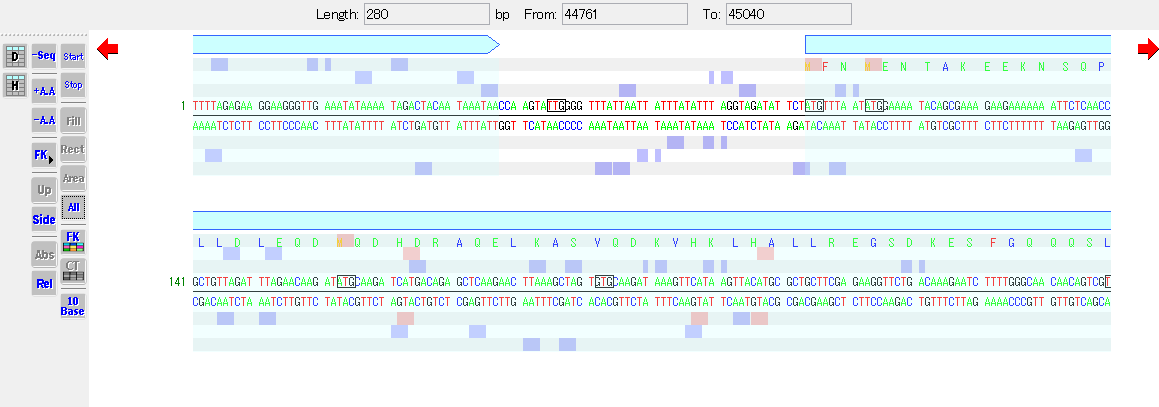
Highlight the area corresponding to the feature
Enclose the base sequence region corresponding to the feature in a rectangle
- Turn on the Rect button in the toolbox to enclose the sequence region corresponding to the feature in a rectangle.
- At this time, the changeable start codon on the base sequence changes to a highlighted display instead of a rectangular display.
Shaded display of the entire area corresponding to the feature
- Select the All button in the toolbox to shade the area corresponding to the feature.
- The shaded color is a transparent version of the color assigned to the feature key.
- At this time, the changeable start codon on the base sequence is surrounded by a rectangle.
Change feature display color
- If you turn on the FK color button in the toolbox, it will be drawn in the color assigned to that feature key.
- How to set the FK color.
- If you turn on the CT color button in the toolbox, it will be displayed in the category color registered for that feature.
- Category colors are set for each annotation progress stage.
- How to set the category color.
Insert a blank character every 10bp of the base sequence
- When the 10Base button in the toolbox is turned on, a blank is inserted in the base sequence display for 10 bases.
- This is a display only, no blanks are inserted in the actual sequence data.
Operations that can be performed on the base sequence
Changed the start codon position of the CDS feature
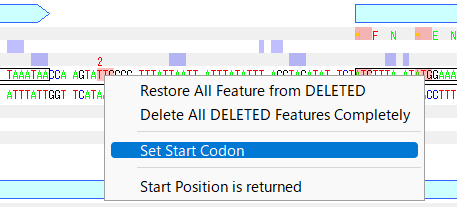
If you want to change the start codon position of the displayed CDS feature to another codon, do the following:
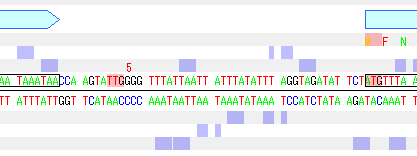
- Right-click the mouse at the change destination start codon position on the base sequence.
- A pop-up menu will appear, select Set Start Codon.
- A confirmation message is displayed.
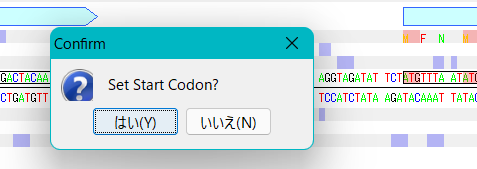
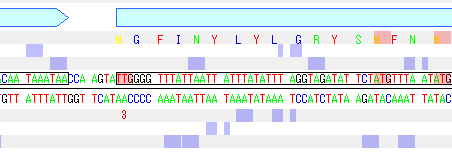
- Answering "yes" will change the start codon position for that CDS feature.
Registration of new features
- Drag the mouse on the base sequence and select the area you want to register as a new feature.
- Right-click on the selected area.
- A pop-up menu is displayed.
- Click Save as (New) Feature.
- The Feature Registration Dialog is displayed.
- The Feature Registration Dialog has the same functionality as the Description Window.
- Enter the required information and click the Apply button.
- A new Feature will be registered in that area.
PCR primer design
You can design PCR primers for selected regions.
- Drag the mouse on the base sequence and select the area you want to register as a new feature.
- Right-click on the selected area.
- A pop-up menu is displayed.
- Click PCR Primer Design.
- Please refer to PCR Primer Design for details.
- If you want to register the selected base sequence as PCR Primer as it is, do it on the sequence lane of the main window.
 Dongle License (HW Key)
Dongle License (HW Key) Feature Map
Feature Map Management and Operations of Feature Keys
Management and Operations of Feature Keys Sequence and Data Input and Output
Sequence and Data Input and Output GenBank EMBL Viewer
GenBank EMBL Viewer Sequence Viewer
Sequence Viewer Annotation Viewer
Annotation Viewer Circular Genome Viewer-Designer
Circular Genome Viewer-Designer Plasmid Map Viewer-Designer
Plasmid Map Viewer-Designer Trace Viewer - Editor
Trace Viewer - Editor Phylogenetic Tree Viewer
Phylogenetic Tree Viewer Feature Key Search
Feature Key Search Keyword Search
Keyword Search Pattern Search
Pattern Search Priming Site Search
Priming Site Search Batch Homology Search
Batch Homology Search Restriction Enzyme
Restriction Enzyme Primer Design
Primer Design PCR Reaction
PCR Reaction Ligation
Ligation Fragment Modification
Fragment Modification DNA Content Analysis
DNA Content Analysis Codon Analysis
Codon Analysis ORF Analysis
ORF Analysis Database Management
Database Management Multiple Circular Genome Map
Multiple Circular Genome Map Dot Plot Analysis
Dot Plot Analysis Venn Diagram Analysis
Venn Diagram Analysis Reverse Complement
Reverse Complement Settings
Settings