IMC B2K1 Register CDS Candidate from Frame Lane
- CDS candidates can be registered as new features from the stop lane stop absence region of the frame lane.
- This is not a CDS prediction function. The GT-AG rule is ignored.
Operation
- We display the feature lane and frame lane in the main feature map.
- Right-click on the stop lane stop codon absent area (orange area in the above figure) in the frame lane.
- The description window will be displayed.
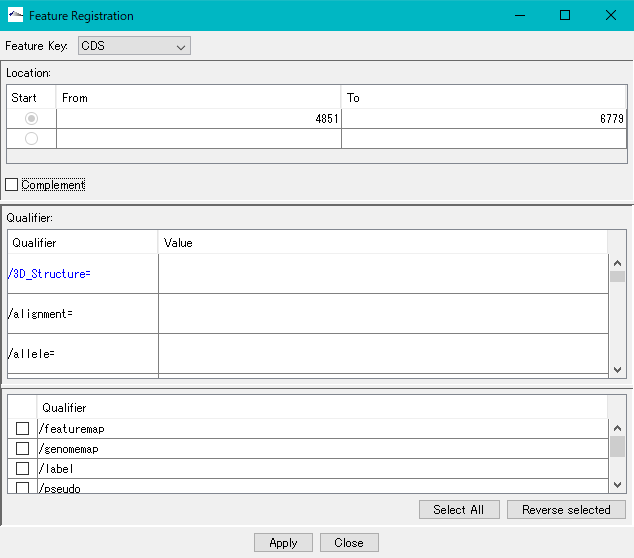
- Click "Apply".
- The CDS corresponding to the selected area is newly registered.
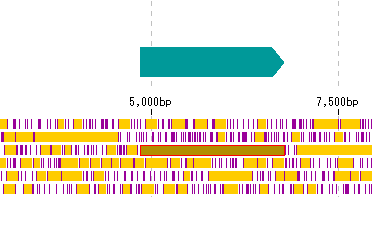
- If CDS display is set in the feature lane, it will be displayed as follows.

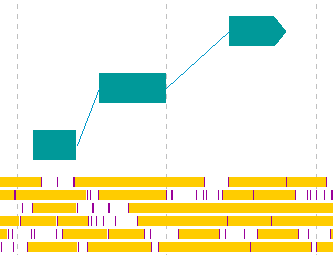
- Register CDS candidates consisting of multiple exons in eukaryotic organisms.
- Display the frame lane in the main feature map.
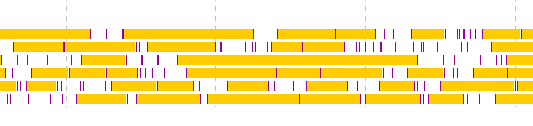
- Click from the first exon candidate to the region immediately before the last exon.
- If you click the last exon candidate before right-clicking the mouse, it will be out of the selection.
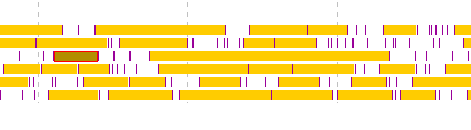
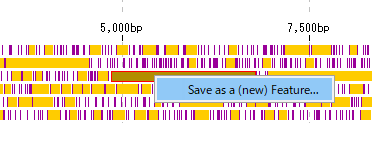
- Right click on the final exon candidate.
- A menu will be displayed and click on "Save as a (new) Feature".
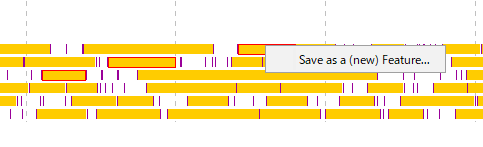
- The description window will be displayed.
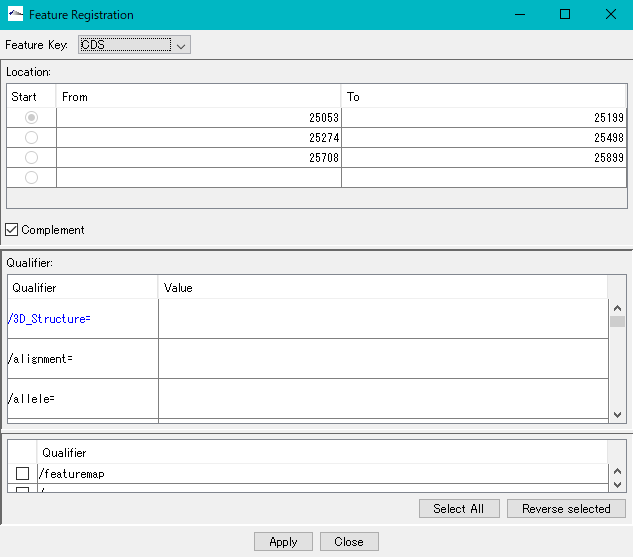
- Position information of the number of areas specified for Location is set.
- Click "Apply".
- A multi-exon CDS will be displayed on the feature lane.

- It is also possible to draw for each frame to which each exon belongs.
 Dongle License (HW Key)
Dongle License (HW Key) Feature Map
Feature Map Management and Operations of Feature Keys
Management and Operations of Feature Keys Sequence and Data Input and Output
Sequence and Data Input and Output GenBank EMBL Viewer
GenBank EMBL Viewer Sequence Viewer
Sequence Viewer Annotation Viewer
Annotation Viewer Circular Genome Viewer-Designer
Circular Genome Viewer-Designer Plasmid Map Viewer-Designer
Plasmid Map Viewer-Designer Trace Viewer - Editor
Trace Viewer - Editor Phylogenetic Tree Viewer
Phylogenetic Tree Viewer Feature Key Search
Feature Key Search Keyword Search
Keyword Search Pattern Search
Pattern Search Priming Site Search
Priming Site Search Batch Homology Search
Batch Homology Search Restriction Enzyme
Restriction Enzyme Primer Design
Primer Design PCR Reaction
PCR Reaction Ligation
Ligation Fragment Modification
Fragment Modification DNA Content Analysis
DNA Content Analysis Codon Analysis
Codon Analysis ORF Analysis
ORF Analysis Database Management
Database Management Multiple Circular Genome Map
Multiple Circular Genome Map Dot Plot Analysis
Dot Plot Analysis Venn Diagram Analysis
Venn Diagram Analysis Reverse Complement
Reverse Complement Settings
Settings