IMC B2DA31 Blast Search by CDS of Selected Area on Feature Lane
Perform a homology search using Blast with the CDS feature included in the selected area of the feature lane of the main feature map as the query sequence.
Homology search results are recorded in the Qualifier of each feature.
These qualifiers can be displayed the features' shapes, colors, labels, etc accordingly.
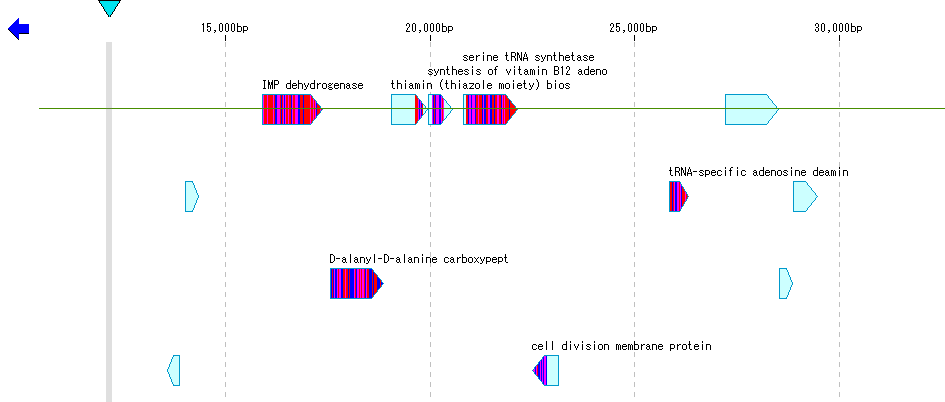
Operation
- The coding region has been identified and the genomic sequence registered as a CDS feature is loaded into the current feature map.
- In the current reference directory, load E_coli.gbk and S_typhimurium.gbk as sample data.
- Drag the mouse and select the area to perform Blast search.
- Right-click on the selection area.
- The menu will be displayed.
- Click "Homology Search by Selected Feature".
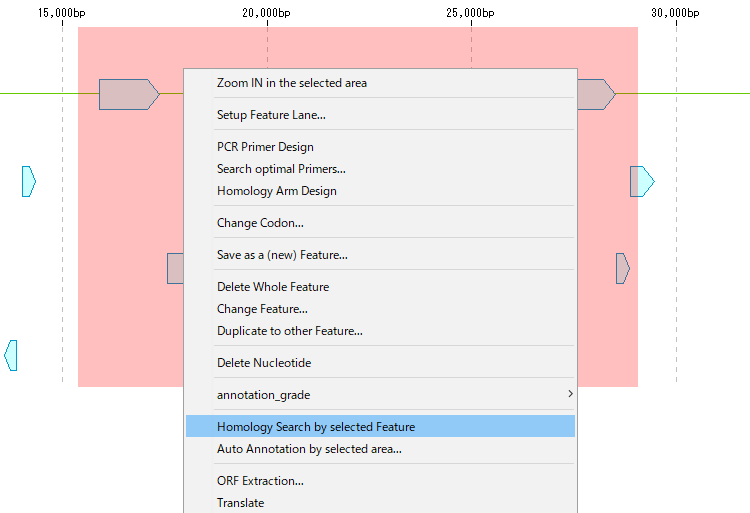
- The "Batch Homology Setting" dialog will be displayed.
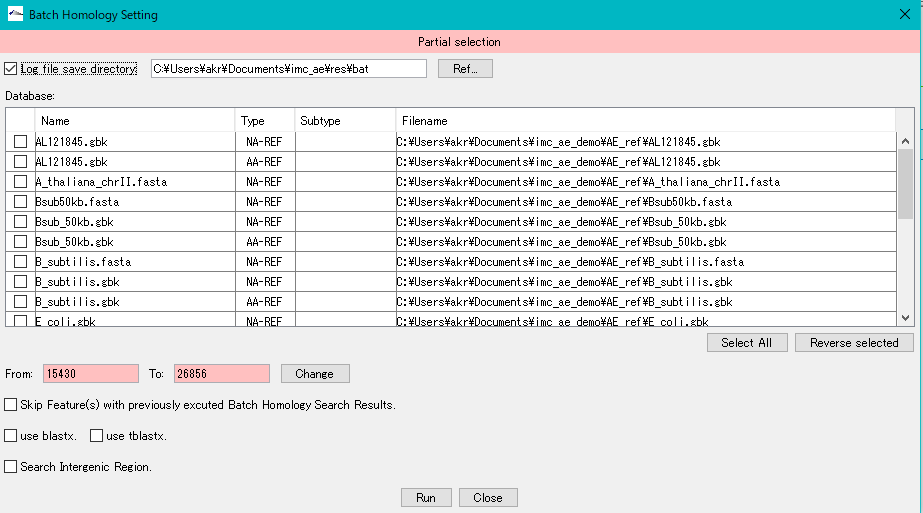
- The nucleotide sequence and amino acid sequence of the genomic sequence loaded in the current reference directory can be selected as a database for Blast search.
- In addition to this, you can create a search database and list it here. (Here)
- Here, as an example, we check the AA-REF of E_coli.gbk and the AA-REF of S_typhimurium.gbk to make it a search database.
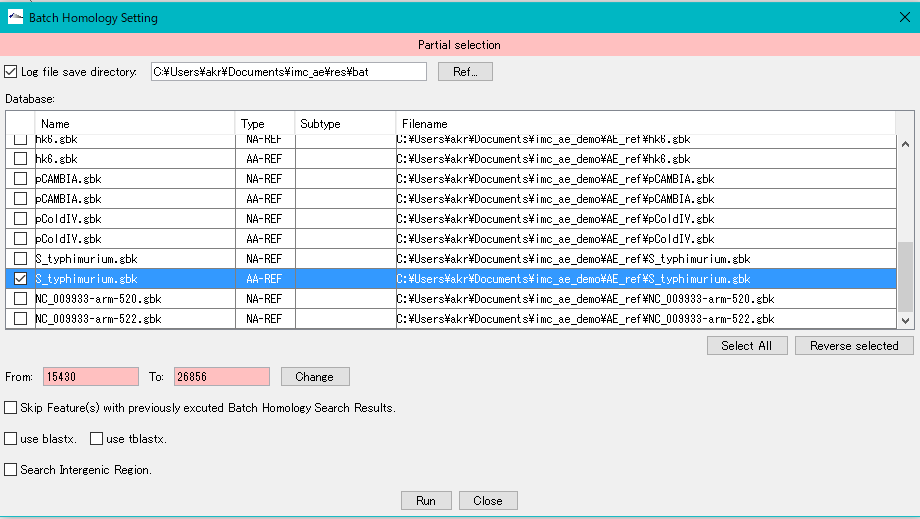
- Click "Run".
- The "Start Homology Search?" Execution confirmation dialog is displayed.
- Click "Show Parameter ...".
- The execution confirmation dialog is expanded and displayed, and Blast search execution parameters are displayed.
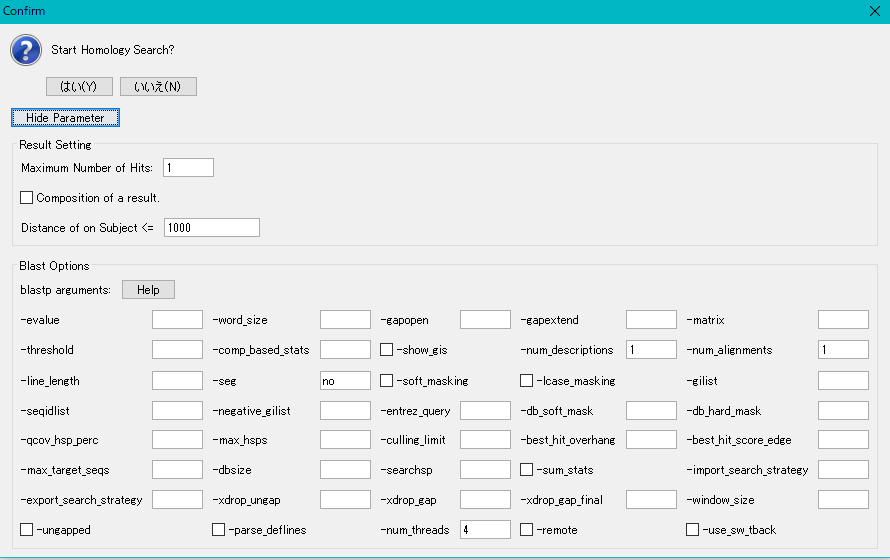
- Enter the maximum number of hits directly as a positive integer in the "Maximum Number of Hits" input field of the Result Setting column.
- This specifies how many search results to save per query.
- Hits greater than the entered number are not saved, but if few hits, etc., this number will save less hits.
- We will set this to 5 here.
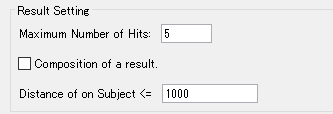
- Click "Yes (Y)".
- Blast search execution starts.
- A progress message is displayed during execution.
- When the search is completed, the search result dialog is displayed.
- You can execute manual annotation from this dialog (explanation is here).
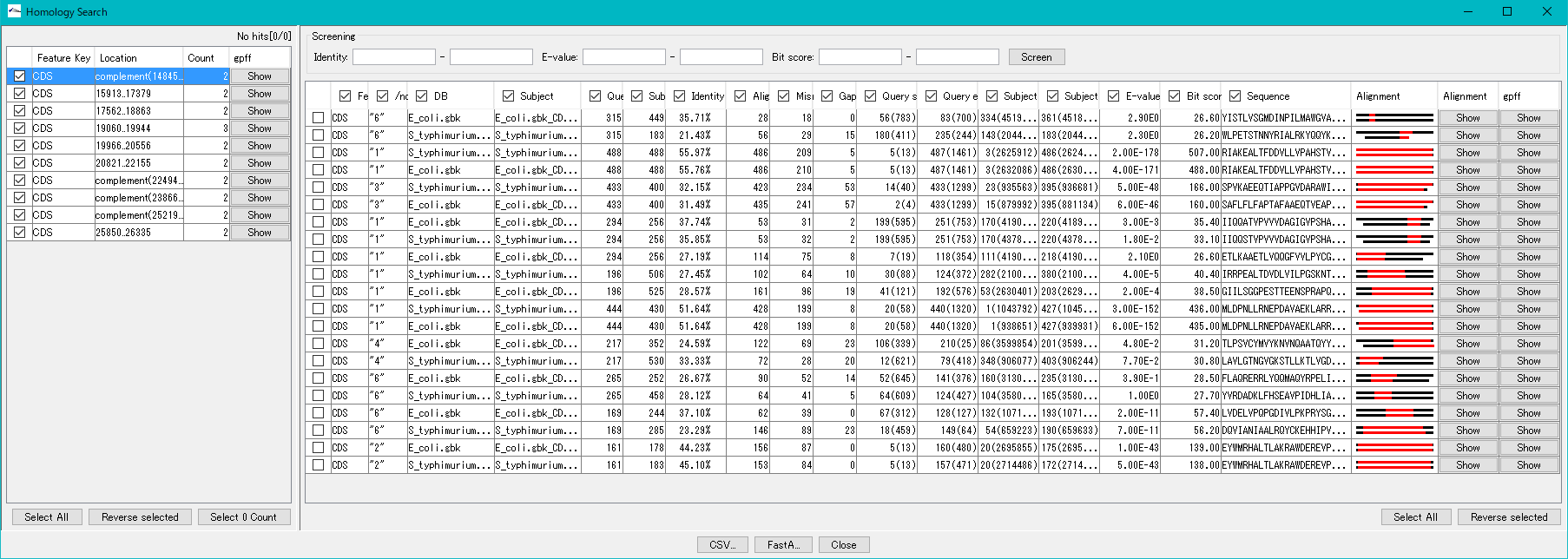
- If "Auto Copy" is set, the top hit qualifier is automatically registered in the query CDS.
- The explanation of "Auto Copy" is here.
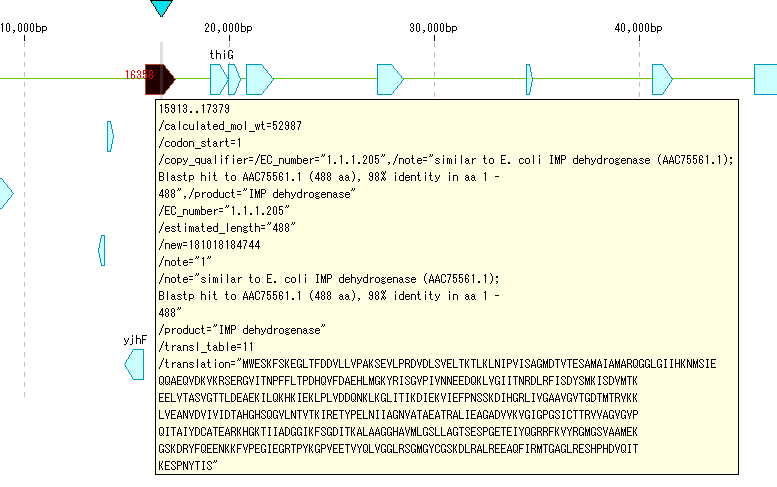
- Manual annotation is available from the search result dialog. Manual annotation is here.
- You will see the results posted automatically.
- Turn on the label button from Toolbox. A label appears above the CDS feature.
- In the case below, gene is labeled.
- Turn off the label button.
- The label above the CDS feature disappears.
- If the label does not disappear even if you turn off the label button, the label is "Permanent Label".
- To cancel this, you need to change that setting from the Feature Map tab pane of the Feature Setting dialog.
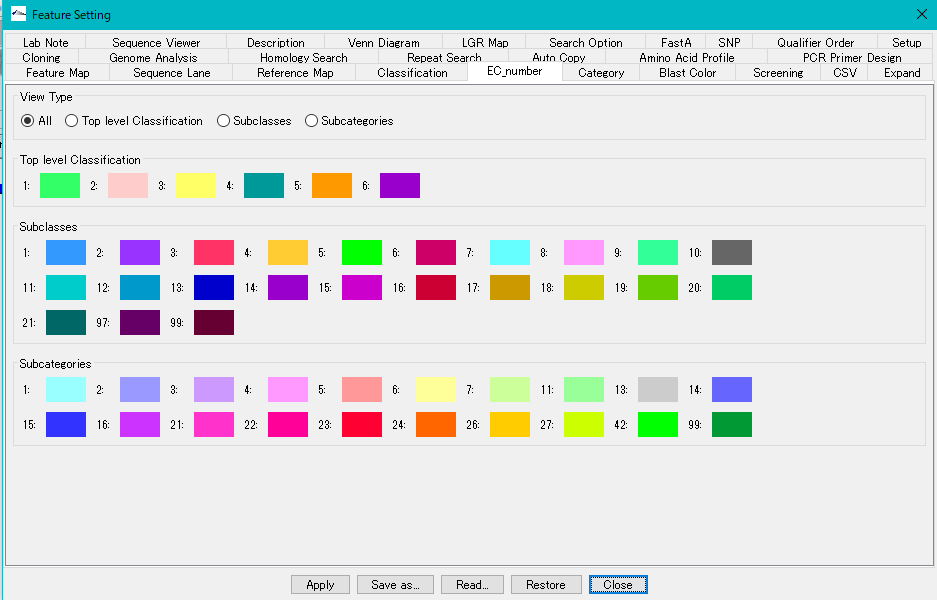
- Click "EC" button from Toolbox.
- The CDS to which the EC number was posted is displayed in the color assigned to the EC number.
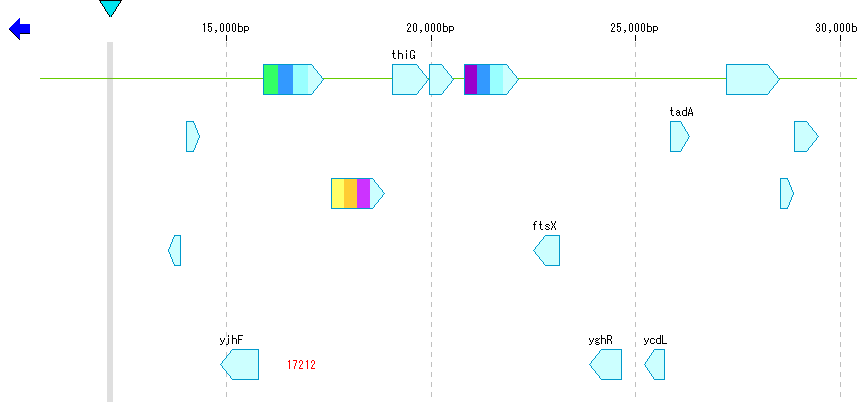
- You can change this color setting in the "EC_number" tab pane.
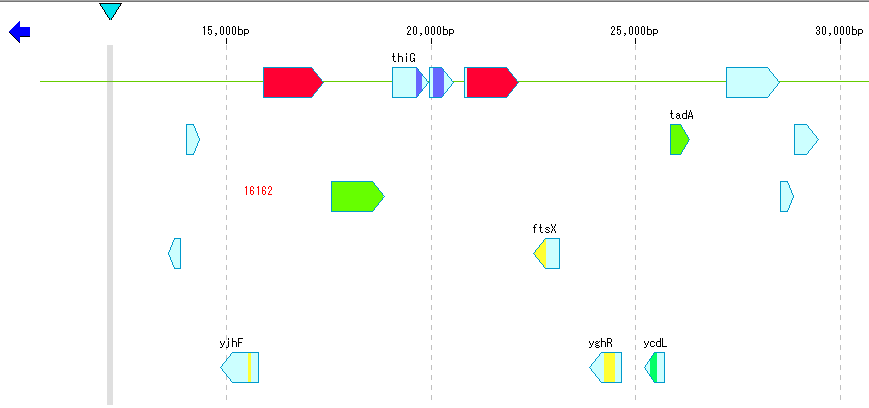
- Click "BL" button from ToolBox.
- The hit CDS feature will be displayed in Blast's score color.
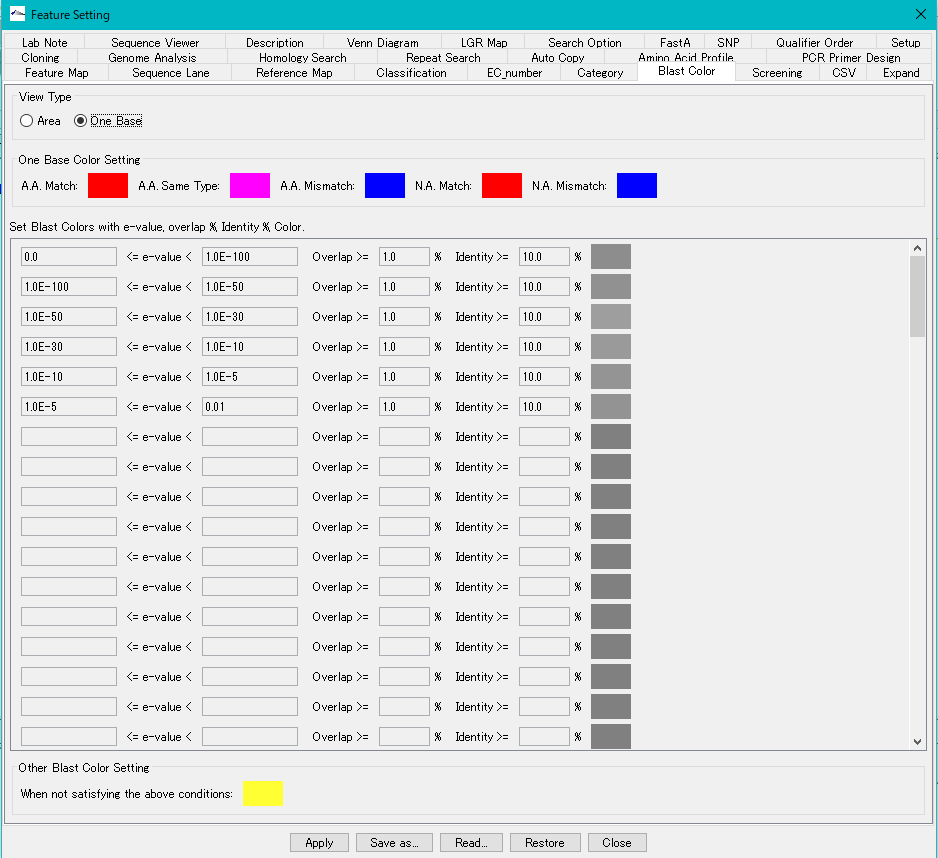
- You can change this color setting in the "Blast Color" tab pane.
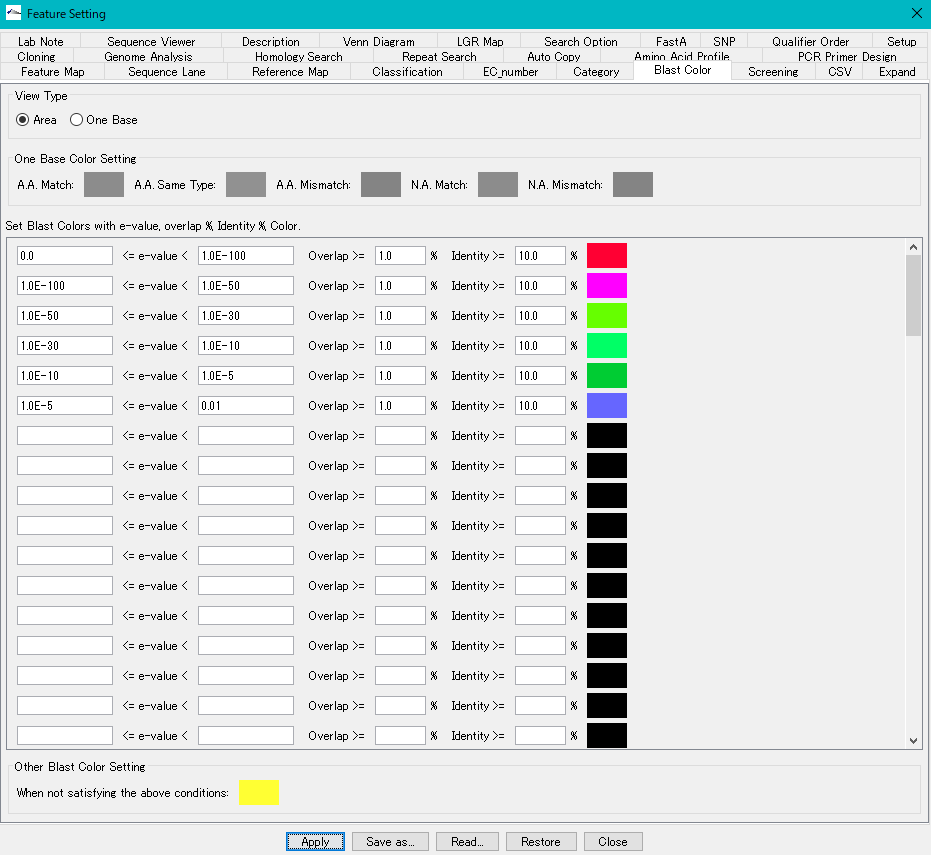
- Blast score color is displayed by hit area, but color display in amino acid residue unit or base unit is also possible.
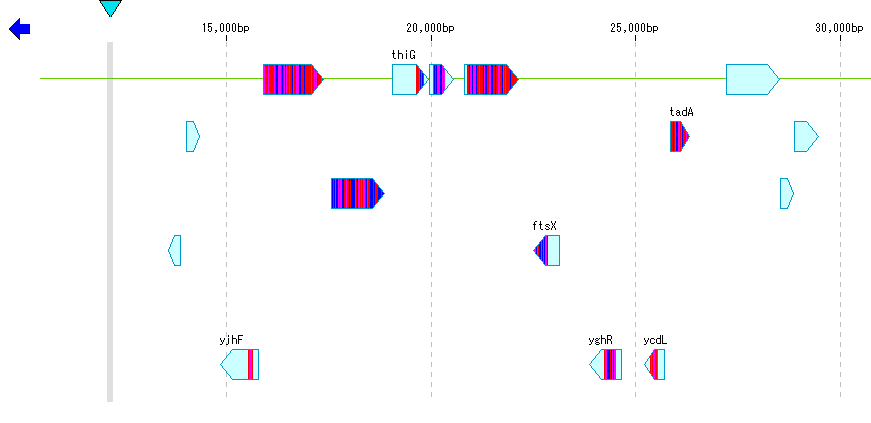
- You can change this color setting in the "Blast Color" tab pane.
- To change the Qualifier used as a label, change it from "Feature Map".
- Change "gene" to "product".
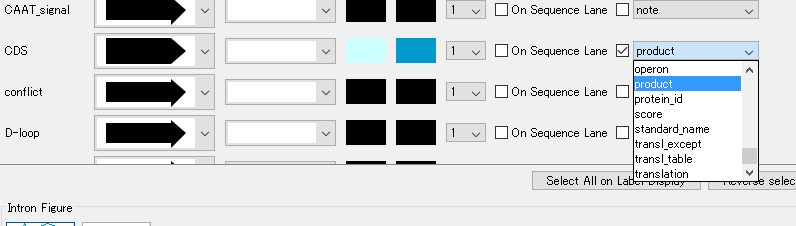
- As a label, the CDS feature's Qualifier "product" is used.

 Dongle License (HW Key)
Dongle License (HW Key) Feature Map
Feature Map Management and Operations of Feature Keys
Management and Operations of Feature Keys Sequence and Data Input and Output
Sequence and Data Input and Output GenBank EMBL Viewer
GenBank EMBL Viewer Sequence Viewer
Sequence Viewer Annotation Viewer
Annotation Viewer Circular Genome Viewer-Designer
Circular Genome Viewer-Designer Plasmid Map Viewer-Designer
Plasmid Map Viewer-Designer Trace Viewer - Editor
Trace Viewer - Editor Phylogenetic Tree Viewer
Phylogenetic Tree Viewer Feature Key Search
Feature Key Search Keyword Search
Keyword Search Pattern Search
Pattern Search Priming Site Search
Priming Site Search Batch Homology Search
Batch Homology Search Restriction Enzyme
Restriction Enzyme Primer Design
Primer Design PCR Reaction
PCR Reaction Ligation
Ligation Fragment Modification
Fragment Modification DNA Content Analysis
DNA Content Analysis Codon Analysis
Codon Analysis ORF Analysis
ORF Analysis Database Management
Database Management Multiple Circular Genome Map
Multiple Circular Genome Map Dot Plot Analysis
Dot Plot Analysis Venn Diagram Analysis
Venn Diagram Analysis Reverse Complement
Reverse Complement Settings
Settings