IMC i01B Find Restriction Enzyme Recognition Sites in Genome Sequence
Search the recognition sites of the specified restriction enzyme (multiple or all can be specified) from the genomic sequence displayed in the main feature map.
Operation flow
- Load genome sequence
- Selection of genome sequence
- Selection of restriction enzyme
- Detection of recognition site
- Feature map display of recognition site
Implementation Edition
EE![]() , SE
, SE![]() , GE
, GE![]() , AE
, AE![]() , DS
, DS![]() , GT
, GT![]()
Operation
- We will explain using the sample genome sequence here.
- Taking the sample genome sequence (Bsub 50 Kbp gbk) as an examble, Click to make it the current genome sequence.
- The sample genome sequence is displayed in the feature map.
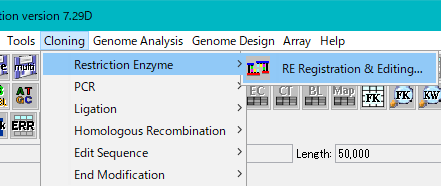
- From the menu, select "Cloning -> Restriction Enzyme -> RE Registration & Editing".
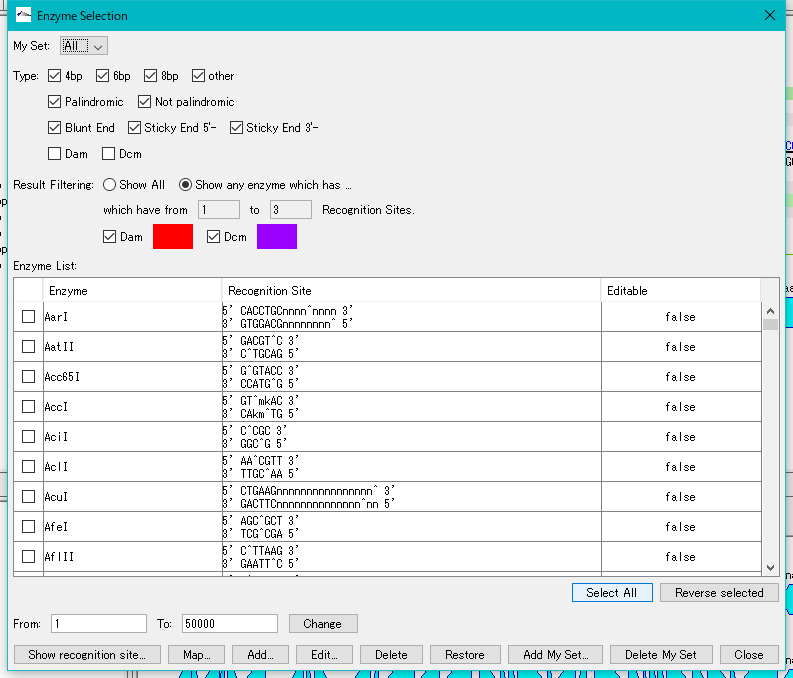
- The "Enzyme Selection" dialog is displayed.
- Check the restriction enzyme to be used (multiple selection or all selection possible). In this case, click "Select All" and check all restriction enzymes.
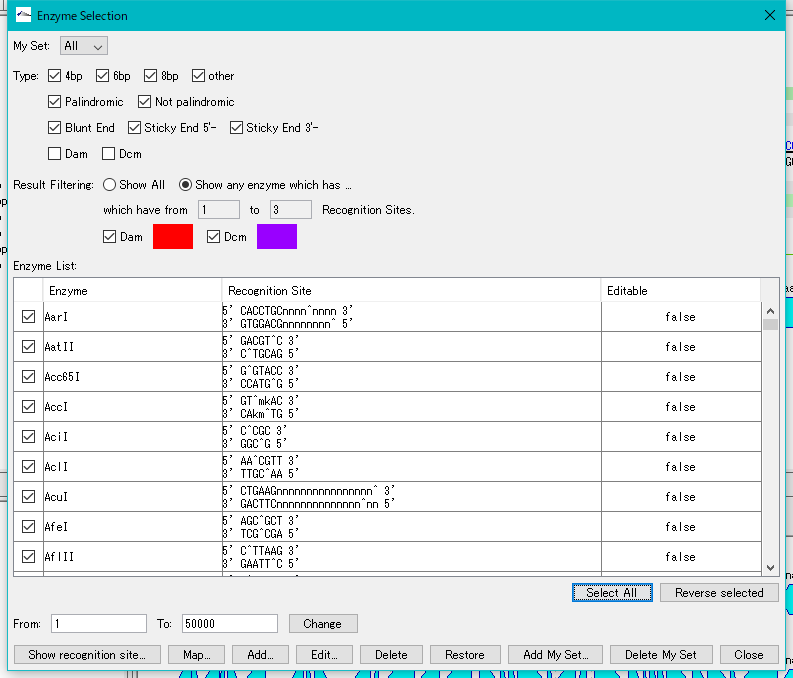
- Click "Show Recognition Site ...".
- The "Recognition Site" dialog is displayed, and the number of restriction sites by restriction enzyme number is listed in the left panel.
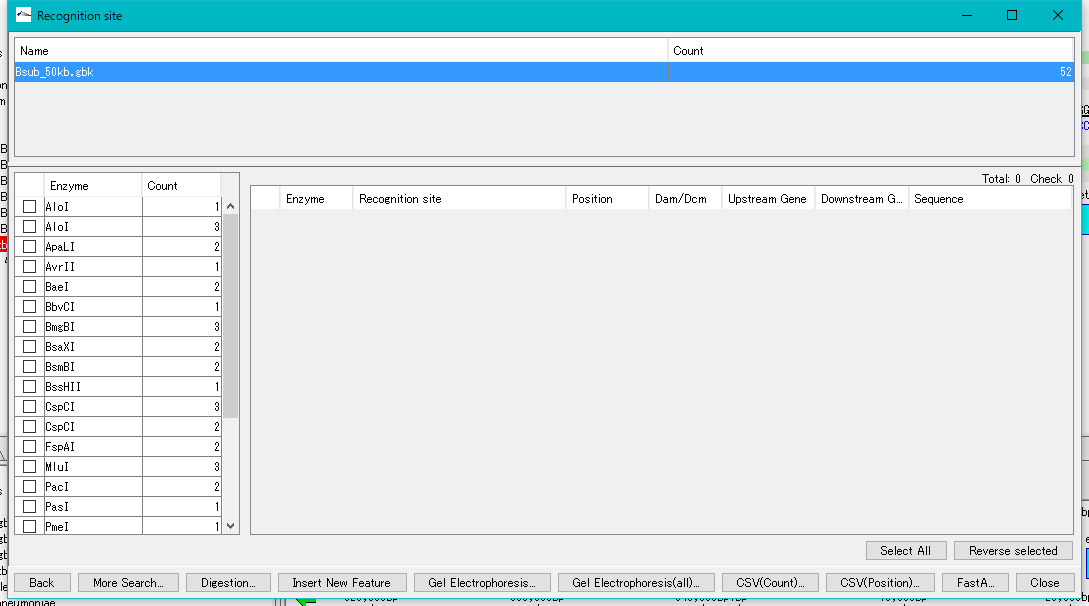
- Restriction enzymes with one or more restriction enzyme recognition sites are displayed on the list, and restriction enzymes that have no restriction site are not displayed.
- Check the checkbox to the left of the restriction enzyme (multiple and all designation possible).
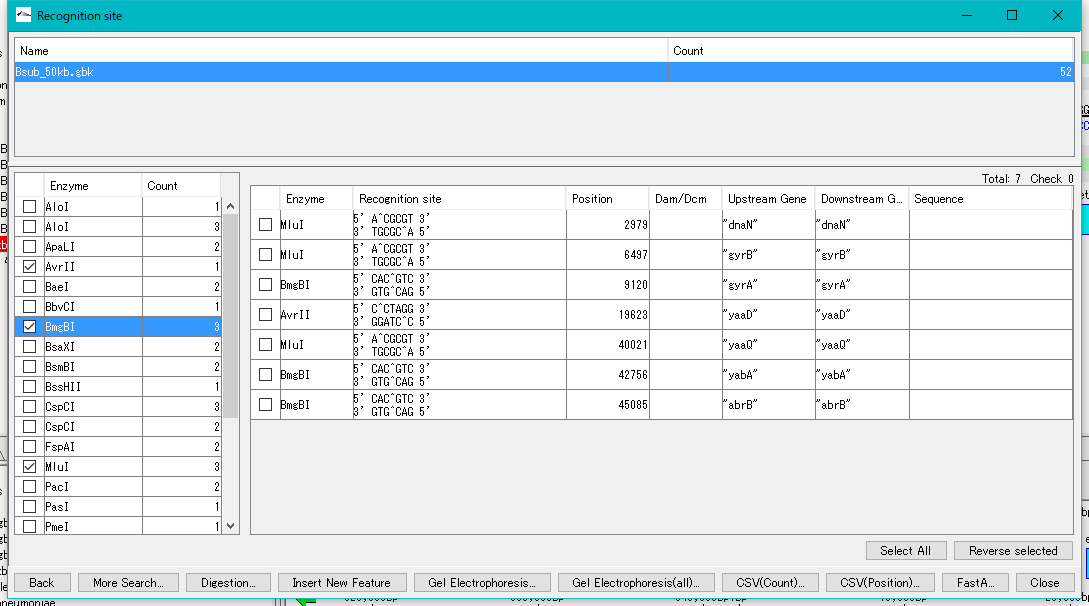
- A list of recognition sites for the selected enzymes is displayed in the right pane.
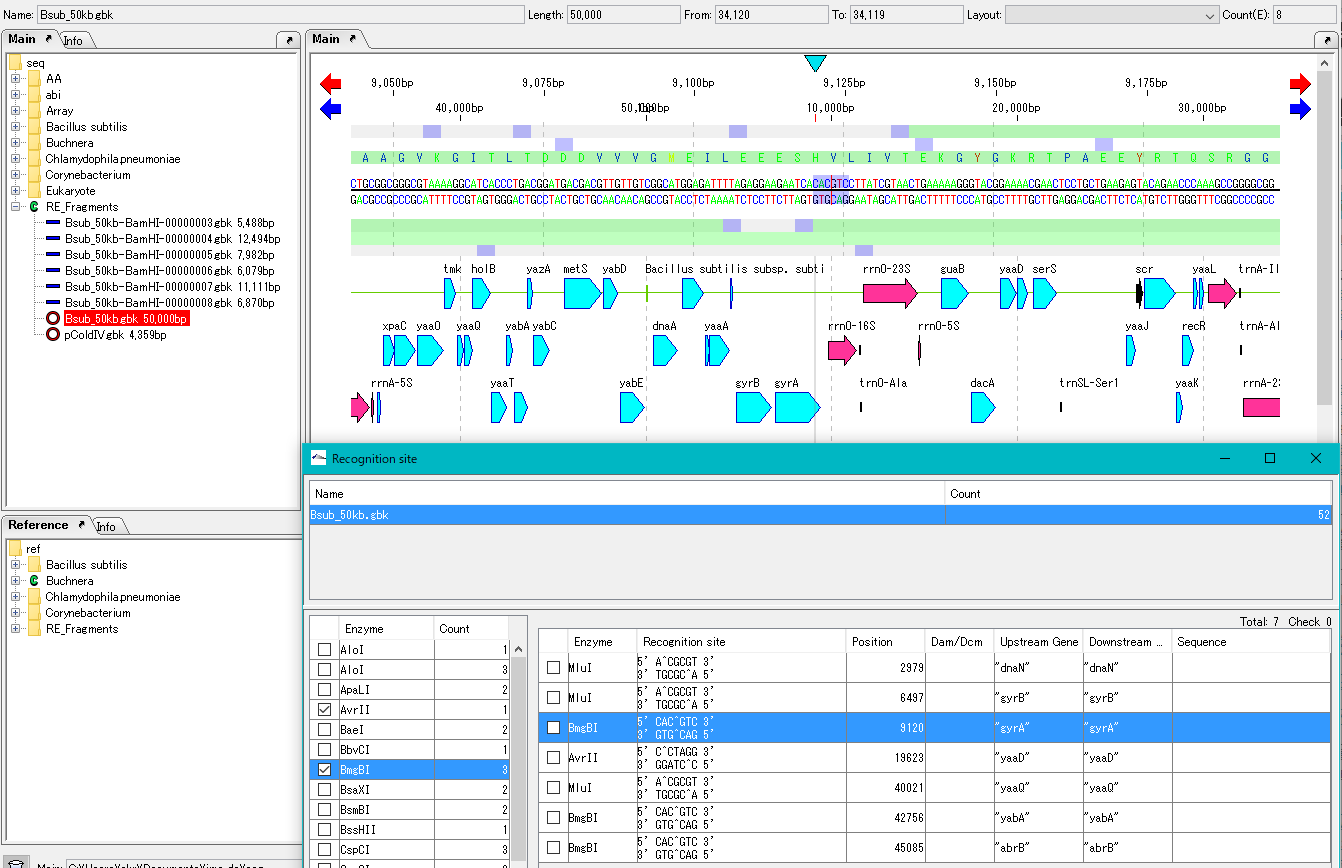
- Click one recognition row in the list on the right panel. The main feature map will automatically scroll and the restriction part will be displayed.
 Dongle License (HW Key)
Dongle License (HW Key) Feature Map
Feature Map Management and Operations of Feature Keys
Management and Operations of Feature Keys Sequence and Data Input and Output
Sequence and Data Input and Output GenBank EMBL Viewer
GenBank EMBL Viewer Sequence Viewer
Sequence Viewer Annotation Viewer
Annotation Viewer Circular Genome Viewer-Designer
Circular Genome Viewer-Designer Plasmid Map Viewer-Designer
Plasmid Map Viewer-Designer Trace Viewer - Editor
Trace Viewer - Editor Phylogenetic Tree Viewer
Phylogenetic Tree Viewer Feature Key Search
Feature Key Search Keyword Search
Keyword Search Pattern Search
Pattern Search Priming Site Search
Priming Site Search Batch Homology Search
Batch Homology Search Restriction Enzyme
Restriction Enzyme Primer Design
Primer Design PCR Reaction
PCR Reaction Ligation
Ligation Fragment Modification
Fragment Modification DNA Content Analysis
DNA Content Analysis Codon Analysis
Codon Analysis ORF Analysis
ORF Analysis Database Management
Database Management Multiple Circular Genome Map
Multiple Circular Genome Map Dot Plot Analysis
Dot Plot Analysis Venn Diagram Analysis
Venn Diagram Analysis Reverse Complement
Reverse Complement Settings
Settings